Finding antibiotic-resistant genes
In this tutorial we will find antibiotic-resistant genes in a bacterial genome.
Get data
- Log in to your Galaxy instance (for example, Galaxy Australia, usegalaxy.org.au).
Use shared data
If you are using Galaxy Australia, you can import the data from a shared data library.
In the top menu bar, go to
- Click on
Data Libraries . - Click on
Galaxy Australia Training Material: Annotation: Microbial Annotation . - Tick the box next to the file.
- Click the
To History button, select As Datasets. - Name a new history and click
Import . - In the top menu bar, click
Analyze Data . - You should now have one file in your current history.
Or, import from the web
Only follow this step if unable to load the data files from shared data, as described above.
- In a new browser tab, go to this webpage:
- Find the file called
contigs.fasta - Right click on file name: select “copy link address”
- In Galaxy, go to
Get Data -> Upload File - Click
Paste/Fetch data - A box will appear: paste in link address
- Click
Start - Click
Close - The file will now appear in the top of your history panel. When it has finished uploading, the file name will be green.
Shorten file name
- Click on the pencil icon next to the file name.
- In the centre Galaxy panel, click in the box under
Name - Shorten the file name.
- Then click
Save
Find antibiotic-resistant genes
We will use the tool called ABRicate to find antibiotic-resistant genes in the (draft) genome.
In the tools panel seach box, type in “abricate”
- Click on
ABRicate - For
Input file choosecontigs.fasta (or the name of your own assembly file.) - Click
Execute .
There is one output file.
- Click on the eye icon to view.
- It should look like this, although likely with a different number of rows.
-
(We have used a miniature genome in this example, so only have a few rows).
-
This shows a table with one line for each antibiotic-resistant gene found, in which contig, at which position, and the % coverage.
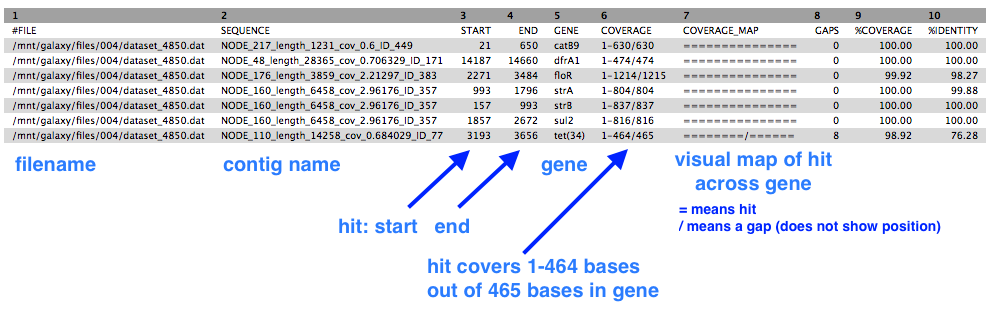
In the output from Abricate, column 5 has the list of the antibiotic-resistant gene names.
-
Some of these may be complete, exact matches, and some may have a gap/mutation in their sequence which can affect whether that protein is actually expressed.
-
To find out more about what type of AMR genes these are, you can search Genbank with the gene name (e.g. aadD).
See this history in Galaxy
If you want to see this Galaxy history without performing the steps above:
- Log in to Galaxy Australia: https://usegalaxy.org.au/
- Go to
Shared Data - Click
Histories - Click
Completed-Abricate-analysis - Click
Import (at the top right corner) - The analysis should now be showing as your current history.
What’s next?
To use the tutorials on this website:
- ← see the list in the left hand panel
- ↖ or, click the menu button (three horizontal bars) in the top left of the page
You can find more tutorials at the Galaxy Training Network: